XIE/Enzyme:2025/Dec: Difference between revisions
(→12.10) |
(→12.10) |
||
| Line 61: | Line 61: | ||
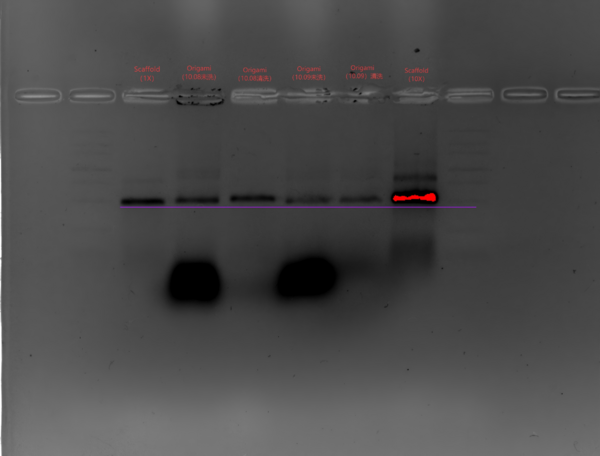
#'''Marker、Scaffold(1X)、Origami(10.08-未洗)、Origami(10.08—清洗)、Origami(10.09-未洗)、Origami(10.09-清洗)、Scaffold(10X)、Marker''' | #'''Marker、Scaffold(1X)、Origami(10.08-未洗)、Origami(10.08—清洗)、Origami(10.09-未洗)、Origami(10.09-清洗)、Scaffold(10X)、Marker''' | ||
[[FIle:Origami(NO)_25_12_10.png|thumb|center|600px|Origami(NO)]] | [[FIle:Origami(NO)_25_12_10.png|thumb|center|600px|Origami(NO)]] | ||
== 12.11 == | |||
*<big>'''TEM'''</big> | |||
=== Negative staining (uranyl acetate) === | |||
*'''Grid activation''': glow‑discharge copper grids for 30 s. | |||
*'''Sample staining''' | |||
#apply 4.5 µL of sample onto the grid. | |||
#Blot off the sample, add buffer to the grid, then blot off the buffer. | |||
#Prepare parafilm and place three drops of uranyl acetate (pH adjusted) on the parafilm. | |||
#Quickly transfer the grid into a staining droplet, blot off excess, and incubate for 30 s. | |||
#Air‑dry the grid. | |||
Revision as of 11:48, 12 December 2025
12.08
- HR-DNA Origami(NO) Folding
- Materials
| Material | Volume(100 μL) |
|---|---|
| Staples (500 nM) | 25 μL |
| P8064 (150 nM) | 10 μL |
| 10X PBS | 10 μL |
| H2O | 55 μL |
- Program folding in PCR thermocycler : The mixed sample was put on a thermal ramp starting with a rapid heat denaturation at 80 °C for 5 min followed by cooling from 80 °C to 60 °C over 20 min, then slow cooling from 60 °C to 24 °C over 14 h.
- Purification (12.09): Reserve a 10 µL aliquot. Add the remaining 90 µL to a 100‑kDa MWCO centrifugal filter unit and dilute to 500 µL with 1× PBS. Centrifuge at 8,000 g for 1.5 min and discard the filtrate . Add UP Water to 500 ul to the filter unit and repeat the centrifugation; perform this wash step three times. After the final wash, adjust the volume in the filter unit to 100 µL with UP Water, invert the filter unit into a fresh collection tube, and centrifuge at 8,000 g for 1.5 min to collect the concentrate.
12.09
- HR-DNA Origami(NO) Folding
- Materials
| Material | Volume(100 μL) |
|---|---|
| Staples (500 nM) | 25 μL |
| P8064 (150 nM) | 10 μL |
| 10X PBS | 10 μL |
| H2O | 55 μL |
- Program folding in PCR thermocycler : The mixed sample was put on a thermal ramp starting with a rapid heat denaturation at 80 °C for 5 min followed by cooling from 80 °C to 60 °C over 20 min, then slow cooling from 60 °C to 24 °C over 14 h.
- Purification (12.10): Reserve a 10 µL aliquot. Add the remaining 90 µL to a 100‑kDa MWCO centrifugal filter unit and dilute to 500 µL with 1× PBS. Centrifuge at 8,000 g for 1.5 min and discard the filtrate . Add 1X PBS to 500 ul to the filter unit and repeat the centrifugation; perform this wash step three times. After the final wash, adjust the volume in the filter unit to 100 µL with 1X PBS, invert the filter unit into a fresh collection tube, and centrifuge at 8,000 g for 1.5 min to collect the concentrate.
12.10
- Agarose Gel Electrophoresis
- Gel Preparation
- Recipe: 2.4 g agarose in 120 mL 0.5× TBE
- Heat in a microwave to dissolve, cool slightly, adjust volume with 0.5× TBE to 120 ul, then add 1.2 mL of 1 M MgCl2 and 12 µL GoldView DNA stain.
- Insert comb and waite for the gel to cool and solidify.
- Sample preparation
- Origami (15 nM): sample:loading buffer = 5:1.
- Scaffold: take 1 µL scaffold, dilute to 10 µL (final concentration 15 nM); scaffold:loading buffer = 5:1.
- Electrophoresis
- Run at 90 V for 120 min in 0.5× TBE with 10 mM MgCl2
- Marker、Scaffold(1X)、Origami(10.08-未洗)、Origami(10.08—清洗)、Origami(10.09-未洗)、Origami(10.09-清洗)、Scaffold(10X)、Marker
12.11
- TEM
Negative staining (uranyl acetate)
- Grid activation: glow‑discharge copper grids for 30 s.
- Sample staining
- apply 4.5 µL of sample onto the grid.
- Blot off the sample, add buffer to the grid, then blot off the buffer.
- Prepare parafilm and place three drops of uranyl acetate (pH adjusted) on the parafilm.
- Quickly transfer the grid into a staining droplet, blot off excess, and incubate for 30 s.
- Air‑dry the grid.